arviz.plot_ess#
- arviz.plot_ess(idata, var_names=None, filter_vars=None, kind='local', relative=False, coords=None, figsize=None, grid=None, textsize=None, rug=False, rug_kind='diverging', n_points=20, extra_methods=False, min_ess=400, labeller=None, ax=None, extra_kwargs=None, text_kwargs=None, hline_kwargs=None, rug_kwargs=None, backend=None, backend_kwargs=None, show=None, **kwargs)[source]#
Generate quantile, local, or evolution ESS plots.
The local and the quantile ESS plots are recommended for checking that there are enough samples for all the explored regions of the parameter space. Checking local and quantile ESS is particularly relevant when working with HDI intervals as opposed to ESS bulk, which is suitable for point estimates.
- Parameters:
- idata
InferenceData Any object that can be converted to an
arviz.InferenceDataobject Refer to documentation ofarviz.convert_to_dataset()for details.- var_names
listofstr, optional Variables to be plotted. Prefix the variables by
~when you want to exclude them from the plot. See this section for usage examples.- filter_vars{
None, “like”, “regex”}, defaultNone If
None(default), interpretvar_namesas the real variables names. If “like”, interpretvar_namesas substrings of the real variables names. If “regex”, interpretvar_namesas regular expressions on the real variables names. See this section for usage examples.- kind{“local”, “quantile”, “evolution”}, default “local”
Specify the kind of plot:
The
kind="local"argument generates the ESS’ local efficiency for estimating quantiles of a desired posterior.The
kind="quantile"argument generates the ESS’ local efficiency for estimating small-interval probability of a desired posterior.The
kind="evolution"argument generates the estimated ESS’ with incrised number of iterations of a desired posterior.
- relativebool, default
False Show relative ess in plot
ress = ess / N.- coords
dict, optional Coordinates of
var_namesto be plotted. Passed toxarray.Dataset.sel(). See this section for usage examples.- grid
tuple, optional Number of rows and columns. By default, the rows and columns are automatically inferred. See this section for usage examples.
- figsize(
float,float), optional Figure size. If
Noneit will be defined automatically.- textsize
float, optional Text size scaling factor for labels, titles and lines. If
Noneit will be autoscaled based onfigsize.- rugbool, default
False Add a rug plot for a specific subset of values.
- rug_kind
str, default “diverging” Variable in sample stats to use as rug mask. Must be a boolean variable.
- n_points
int, default 20 Number of points for which to plot their quantile/local ess or number of subsets in the evolution plot.
- extra_methodsbool, default
False Plot mean and sd ESS as horizontal lines. Not taken into account if
kind = 'evolution'.- min_ess
int, default 400 Minimum number of ESS desired. If
relative=Truethe line is plotted atmin_ess / n_samplesfor local and quantile kinds and as a curve following themin_ess / ndependency in evolution kind.- labellerLabeller, optional
Class providing the method
make_label_vertto generate the labels in the plot titles. Read the Label guide for more details and usage examples.- ax2D array_like of
matplotlib AxesorBokeh Figure, optional A 2D array of locations into which to plot the densities. If not supplied, ArviZ will create its own array of plot areas (and return it).
- extra_kwargs
dict, optional If evolution plot,
extra_kwargsis used to plot ess tail and differentiate it from ess bulk. Otherwise, passed to extra methods lines.- text_kwargs
dict, optional Only taken into account when
extra_methods=True. kwargs passed to ax.annotate for extra methods lines labels. It accepts the additional keyxto setxy=(text_kwargs["x"], mcse)- hline_kwargs
dict, optional kwargs passed to
axhline()or toSpandepending on the backend for the horizontal minimum ESS line. For relative ess evolution plots the kwargs are passed toplot()or toline- rug_kwargs
dict kwargs passed to rug plot.
- backend{“matplotlib”, “bokeh”}, default “matplotlib”
Select plotting backend.
- backend_kwargs
dict, optional These are kwargs specific to the backend being used, passed to
matplotlib.pyplot.subplots()orbokeh.plotting.figure. For additional documentation check the plotting method of the backend.- showbool, optional
Call backend show function.
- **kwargs
Passed as-is to
matplotlib.axes.Axes.hist()ormatplotlib.axes.Axes.plot()function depending on the value ofkind.
- idata
- Returns:
- axes
matplotlib AxesorBokeh Figure
- axes
See also
essCalculate estimate of the effective sample size.
References
[1]Vehtari et al. (2021). Rank-normalization, folding, and localization: An improved Rhat for assessing convergence of MCMC. Bayesian analysis, 16(2):667-718.
Examples
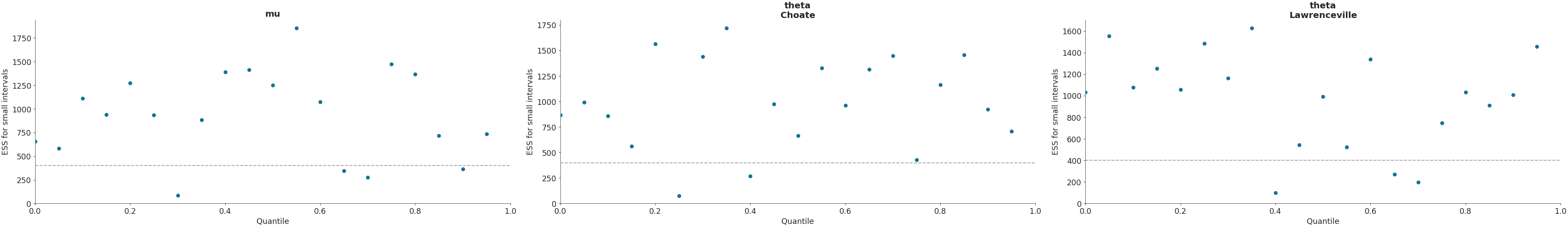
Plot local ESS.
>>> import arviz as az >>> idata = az.load_arviz_data("centered_eight") >>> coords = {"school": ["Choate", "Lawrenceville"]} >>> az.plot_ess( ... idata, kind="local", var_names=["mu", "theta"], coords=coords ... )
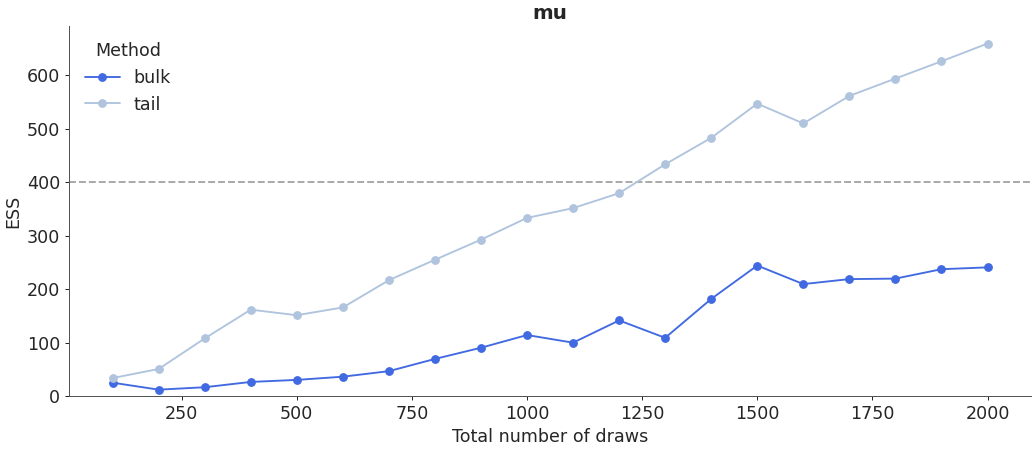
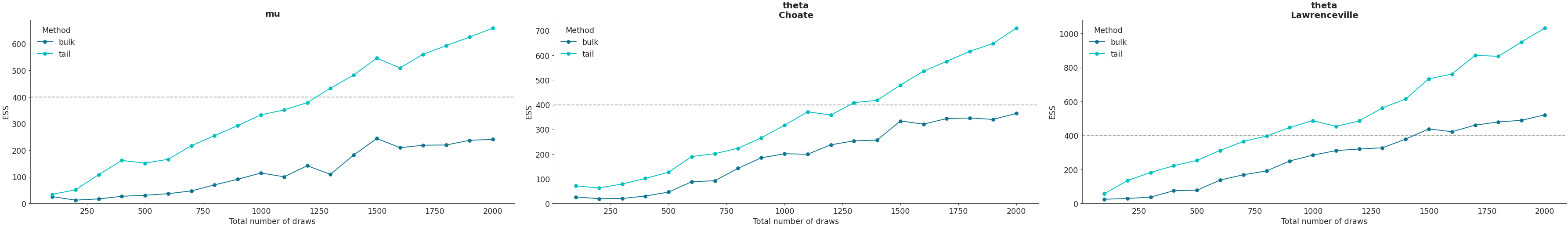
Plot ESS evolution as the number of samples increase. When the model is converging properly, both lines in this plot should be roughly linear.
>>> az.plot_ess( ... idata, kind="evolution", var_names=["mu", "theta"], coords=coords ... )
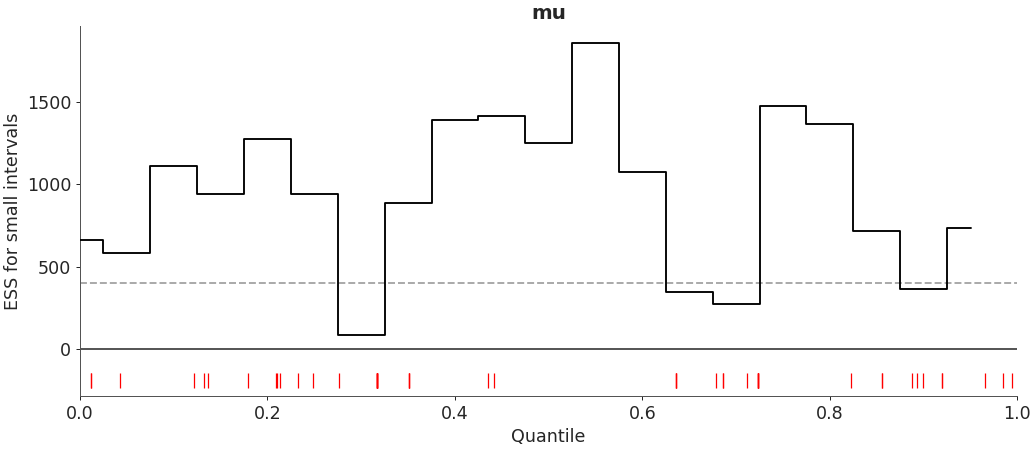
Customize local ESS plot to look like reference paper.
>>> az.plot_ess( ... idata, kind="local", var_names=["mu"], drawstyle="steps-mid", color="k", ... linestyle="-", marker=None, rug=True, rug_kwargs={"color": "r"} ... )
Customize ESS evolution plot to look like reference paper.
>>> extra_kwargs = {"color": "lightsteelblue"} >>> az.plot_ess( ... idata, kind="evolution", var_names=["mu"], ... color="royalblue", extra_kwargs=extra_kwargs ... )